
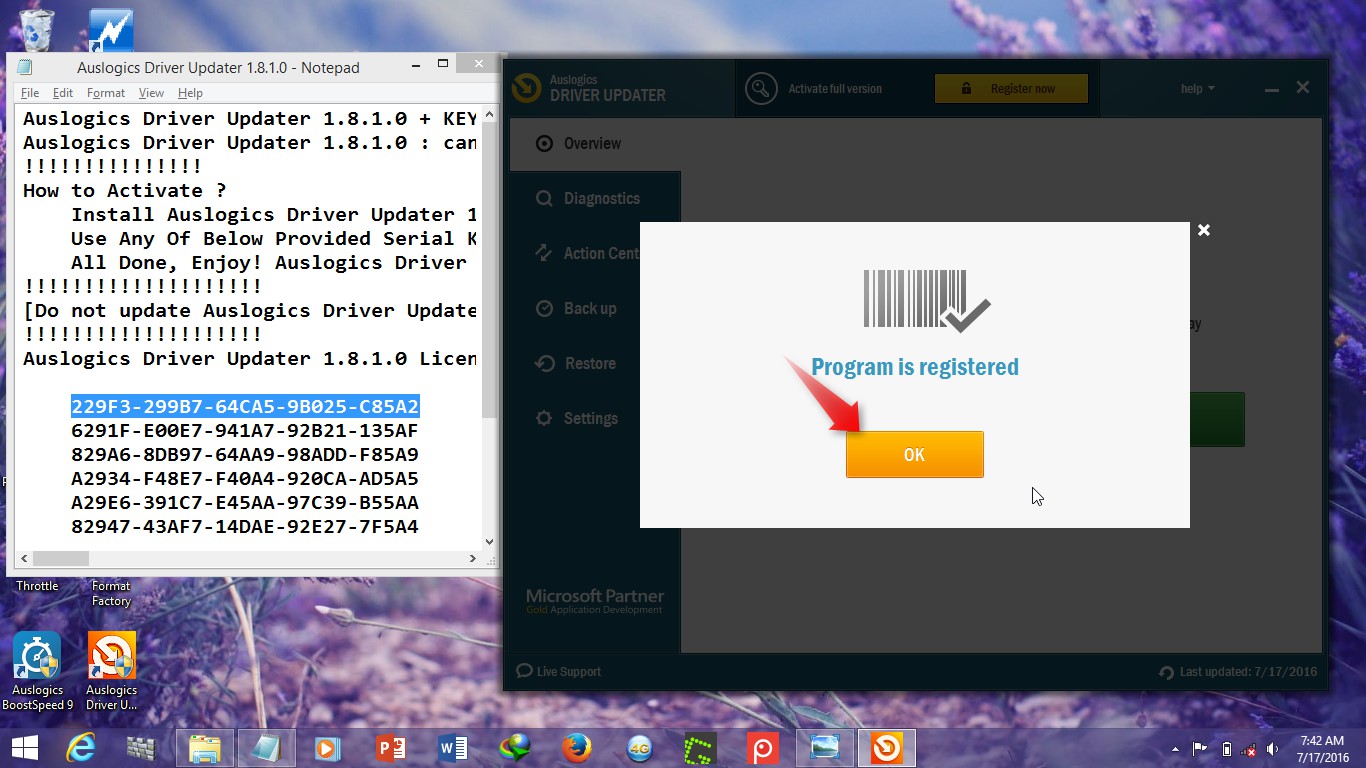
- Bitplane Imaris V7 2 3 Cracked Eggs Mod
- 2/3 Symbol
- Bitplane Imaris V7 2 3 Cracked Eggs
- Imaris Bitplane Support
- Bitplane Imaris V7 2 3 Cracked Eggs Mod
- 2/3 As A Decimal
- Bitplane Imaris V7 2 3 Cracked Eggs Game
- If you search for Bitplane Imaris 7.4.2 X86 X64 Crack. Jun 13, 2016 - url=- Bitplane imaris v7 2 3 -cracked (Bitplane imaris v7 2 3 -cracked.zip)/url. Imaris محصولی قدرتمند از Bitplane بوده که به طور تخصصی برای مصورسازی و تفسیر دادههای میکروسکوپ ارائه شده است.
- Crack download software PaleoScan v2020 x64 FracproPT 2019 LandMark 5000dsg10ep4 shipn Leica. Agisoft.PhotoScan.Professional.v1.3.2.4164 x64. Bitplane Imaris v7.4.2.
Bitplane Imaris v7.0.0 x86/x64 195MB Imaris is Biplane s core software module that delivers all the necessary functionality for visualization, segmentation and interpretation of 3D and 4D microscopy datasets. Combining speed, precision and ease-of-use, Imaris provides a complete set of features for working with thre! Cortona3D RapidAuthor 8.1 + RapidDeveloper 2.4 Win64 Digital.Vision.Nucoda.v2015.3.020.Win64 Digital.Vision.Phoenix.v2015.3.020.Win64 IMOLD.V13.SP3 MBend v3.5.148 Atmel Studio v7.0 Carlson Survey Embedded v2016 Geomagic Freeform Plus v2016.0.22 x64 Maplesoft Maple 2016 x64 Aurora FEST3D v2018 Aurora SPARK3D v2018 Leica.LISCAD.v12.0 BendCAM v5.2.
#Title:Bitplane Imaris v7.1.1.Win64-TFT keygen #Tags. VMware Workstation 7.1.3 Build 324285 Final crack IncrediMail Premium 2 6.0.3 Build 4456 crack. Crack software download Datakit Cross Manager v2014.1 Cliosoft SOS v5.31 Linux CHAM. Bitplane Imaris v7.2.3 Samsung Intercept Reviews and Video: scscs3: 0. Found results for Bitplane Imaris crack. Bitplane Imaris 7.2.3 X64. Bitplane Imaris V6.2.0-tft: Bitplane Imaris V7.1.0 Win32-tft.
However, all tracks will be brand new to PSP. Free download font styles characters.
What's New in Imaris 9.2 As imaging technology improves to capture entire specimens at higher resolution over long timescales, biologists are generating larger and longer static and time-lapse images. Imaris 9.2 responds with the ability to identify and analyze millions of cells, nuclei and vesicles within these Big Data images. This latest version brings a further Big Data revolution, enabling the interactive rendering and analysis of millions of Spots. In addition, new editing tools and visualization features for both Spots and Tracks improve the entire analysis workflow saving you valuable time!
What's New in Imaris 9.2 As imaging technology improves to capture entire specimens at higher resolution over long timescales, biologists are generating larger and longer static and time-lapse images. Imaris 9.2 responds with the ability to identify and analyze millions of cells, nuclei and vesicles within these Big Data images. Sony vegas pro 11 serial number.
Adobe media encoder cs6 torrent. • Destination Publishing to Adobe Stock Contributor • Adobe Team Projects support • Support for Color Profiles from After Effects sources • Automatically relinked assets when importing a sequence • Option to maintain playback in Premiere Pro while rendering in Media Encoder • Streamlined bit depth and alpha channel settings (QuickTime only)Export support for ambisonic audio (H.264 only) • Support for Hybrid LOG Gamma (HLG) • Support for HDR in HEVC (H.265) export Export support for Panasonic AVC-Long GOP Op1b.

This latest version brings a further Big Data revolution, enabling the interactive rendering and analysis of millions of Spots. In addition, new editing tools and visualization features for both Spots and Tracks improve the entire analysis workflow saving you valuable time!
Armani_global_images_v7.2.3.0khcmid
What's New in Imaris 9.2 As imaging technology improves to capture entire specimens at higher resolution over long timescales, biologists are generating larger and longer static and time-lapse images. Imaris 9.2 responds with the ability to identify and analyze millions of cells, nuclei and vesicles within these Big Data images. This latest version brings a further Big Data revolution, enabling the interactive rendering and analysis of millions of Spots. In addition, new editing tools and visualization features for both Spots and Tracks improve the entire analysis workflow saving you valuable time!
Welcome to the Imaris 8.3 release notes. Please have a look to the overview of Imaris Release notes for information about prior released features and fixed bugs.
Version Date: May 27, 2016
Membrane Based Cell Boundary Detection
Using Imaris’ Cell component it is now possible to detect cell boundaries based on a membrane signal even if no nucleus signal is available. The following screenshots show examples of this segmentation:
Algorithmically Imaris performs a multi-scale watershed with region merging to achieve this segmentation. Users can interactively adjust the merging thresholds to get an immediate visual effect of the threshold settings. In addition after running the membrane based cell boundary detection users can go to the edit tab of Cells and Merge or Split cells that were not segmented correctly.
Cells Visualization Improved
Improved visualization of Cells on 2D slicers facilitate easier validation of segmentation results. New features include:
Border rendering of cells as white lines
Region rendering of cells with original intensities overlayed by cell color
Cells Color Options
Cells can now be colored by ObjectID to facilitate discrimination of neighbouring cells.
Lineage Tracking Algorithm “Maximum Overlap” for Cells
To track cells that are segmented from a membrane signal the “maximum overlap” tracking algorithm was added to Imaris8.3. This algorithm can track dividing cells to produce lineages.
The algorithm works as follows: For each cell the algorithm searches a mother cell at the previous time point by calculating the spatial overlap with all cells of the previous time point and picking as the mother that cell with which it has the largest overlap. As a sanity check the link to the mother is created only if the overlap exceeds a minimum ratio which is 0.5 by default.
Bitplane Imaris V7 2 3 Cracked Eggs Mod
Reference Frame Component
A Reference Frame is Cartesian Coordinate System that can be positioned and oriented anywhere within an image.
Manual positioning:
- Select reference frame in surpass tree.
- Select Navigate mode
- Press Ctrl while doing 3D rotation or translation (as you would for 3D navigation) to move the reference frame.
Automatic positioning:
- Reference Frames can be created from tracks via the “Correct Drift” button on the track-editor tab of Spots, Surfaces, Cells or Filaments. They are then positioned in such a way that they “follow” the motion of the tracks.
- Reference Frames can be created from the Image Processing menu by choosing ImageAlignment->Set Reference Frames.
Reference Frames Affect Statistics Values
Statistics values of Spots, Cells, Surfaces, Filaments are reported both in the image coordinates and in reference frame coordinates (if the choice of coordinate system has an influence on the values).
For example “Spot Position X” will also be reported in reference frame coordinates as a new statistics value “Spot Position X Reference Frame” when a reference frame exists. The same happens for other positional values and for vectorial values like velocities and for speed values that also depend on the reference frame.
Reference Frame Play Mode
When reference frames are used in time series it is possible to use the “Reference frame play mode” to play through the time series in such a way that the selected reference frame stays still while the time series plays.
If the reference frames are positioned to move with an object this play mode allows to play through the time series in such a way that the “tracked” object stays still.
Tracking in Reference Frame Coordinates
When a reference frame object exists it is possible to choose this reference frame as the coordinate system within which to perform tracking. This is useful when tracking vesicles that move inside a cell which moves itself.
Cells Split Option “One Nucleus per Cell”
Imaris8.3 has two options to perform the split “One Nucleus per Cell”:
Distance From Nucleus - Every pixel within the cells is assigned to the nucleus that it is closest to. All the pixels assigned to the same nucleus make up one cell. This method performs a robust split where borders end up “in the middle” between nuclei.
Cell Channel Intensity - Bright regions of the cell channel around a nucleus are assigned to that nucleus with the effect that the borders between cells end up at low intensity regions. This method tends to fail when two cells are not delimited by a low intensity region.
The “Distance from Nucleus” option is in many cases similar to the previous “Split one Nucleus per Cell” method.
2/3 Symbol
New Statistics Values
Tracks have new statistics values for displacement in the scenario of lineages:
- Displacement Since Previous Division
- Displacement Since Previous Division^2
- Cell Cycle Displacement
- Time Since Track Start
- Velocity Angle X, Velocity Angle Y, Velocity Angle Z
Track Displacement definition for the lineage situation now is the maximum distance of the first position to any of the positions at the last time point of the track. This statistics was in 8.2 not well defined for tracks with multiple objects in the last time point.
Every object has a new statistics value - Distance From Origin.
Vantage 1D Statistical Tests
Vantage1D shows univariate distributions of statistics values for comparison between groups. In previous versions of Imaris it was possible to inspect visually whether distributions differ. Imaris now provides statistical tests to assess differences between distributions. The tests that are reported in the Statistical Tests tab in Vantage1D are the following:
Wilcoxon-Test Tests whether two samples come from continuous distributions with the same medians. F-Test Tests whether two samples come from normal distributions with the same variance.
T-Test Tests whether two samples come from normal distributions with the same means. Kolmogorov-Smirnov-Test Tests whether two samples come from the same populations.
It is possible for users to adjust the confidence level and whether the tests are performed left-tailed, right-tailed or two-tailed.
XT works with new Matlab Component Runtime (MCR) Versions
Imaris works with MCR v7.1 (Matlab R2009a/b) as before and additionally with MCR v8.4 (Matlab R2014b) and it is possible to configure XTensions to use other run Matlab Component Runtimes. Each XTension specifies which MCR it is compiled for in the xml file. The XTensions that are installed with Imaris8.3 are built for MCR v8.4.
Installation of the Matlab Component Runtimes is simplified by starting download directly from Imaris Preferences CustomTools section.
Imaris without Arena

Bitplane Imaris V7 2 3 Cracked Eggs
Due to customer requests we decided to provide a version of Imaris that does not have Arena (and no batch processing capability either). This version is similar to pre-Imaris8 versions in that it can open and save one image at a time. To get this version users can choose to install Imaris without Arena during installation.
Imaris Bitplane Support
Speedups from Parallel Watershed Algorithm
Bitplane Imaris V7 2 3 Cracked Eggs Mod
The watershed algorithm in Imaris runs blockwise and parallel. This produces speedups (compared to previous Imaris versions) for the following steps:
- Surfaces - Split touching Objects (Region Growing)
- Spots - Different Spot Sizes (Region Growing)
- Cells - Split of Nuclei by Seed Points
- Cells - Split Touching Cells
- Cells - Detect Cell Boundary from Cell Membrane
2/3 As A Decimal
Fixed Bugs
Bitplane Imaris V7 2 3 Cracked Eggs Game
| Fixed 99 Bugs (since Imaris 8.2) | |
| 9399 | Drift Correction wrong |
| 9381 | Using an ROI of one time point with Cell and selecting a cell object crashes Imaris |
| 9355 | Possibility to select which MATLAB version to use together with Imaris |
| 9344 | Region Growing Threshold display is not visible if cells rendered on a slicer XY |
| 9338 | Incorrect coloring of track segments and connections in track lineage editor after connecting two objects. |
| 9332 | Imaris shuts down when user deletes a contour surface object |
| 9328 | When RecordingDate value for Leica files is not present Imaris uses '-4713-11-24 00:00:00.000' |
| 9326 | File converter 8.2 crash with OME tif files |
| 9324 | License issues with Imaris 8.2.0 Track vs. LineageTrack |
| 9315 | Track lineage editor in native theme uses white branch connections against a white background |
| 9312 | Wrong formula for velocity in reference manual |
| 9310 | Imaris holds at 'finishing up' on the flash screen when Arena is active |
| 9308 | Opening an OME image causes DrawVolume call to freeze |
| 9305 | Filament No. Dendrites Branches illustration not clear. |
| 9299 | Enabling Nucleus visibility slows Display Adjustment responsiveness |
| 9294 | 'Filaments points tracks' XTension crashes when trying to create the spots object |
| 9292 | Filament branch hierarchy XT does not work in Imaris 8.3 with matlab 2014b |
| 9290 | Automatic threshold for Cell segmentation does not work in batch processing |
| 9280 | Vesicle outside Cell XTension has '1' based CellID (Imaris is zero based) |
| 9258 | Arena Data storage location unable to be moved - file not found error |
| 9244 | File with lots of manually edited objects can't be opened again |
| 9233 | importing IMX file with contour lines crashes Imaris when surface is generated |
| 9231 | Documentation link for Spots Edit Dialog in Spots Wizard wrong |
| 9223 | Automatic caching of image data if data set fits completely into the assigned RAM |
| 9219 | Imaris not using .IMS color table when default colors are selected in Preferences |
| 9208 | Document for the Cell module what negative Vesicle To Nucleus distances mean |
| 9197 | Show Help does not work in Track Editor |
| 9180 | Filament IDs change on editing - causing problems with labels |
| 9174 | Corrupt Statistics display after deleting one channel |
| 9173 | if image is opened in Surpass and file not read completely, deleting the image from Arena will not delete the file on disk |
| 9163 | Description of 'Imaris Administrator - Data Management' is missing |
| 9154 | Imaris quit and will never load .nd2 files (Nikon SIM) |
| 9150 | No channel check boxes are selected in all the Image Processing operations |
| 9142 | Measurement Points Center to Selection does not work if one point is selected |
| 9140 | Deleting multiple assays which contain image data in quick succession will cause Imaris to crash |
| 9138 | Radiobuttons for 'Add / Delete (Cursor intersects with):' are truncated in Edit Spots |
| 9137 | CTRL-S does not store dataset |
| 9129 | Correcting Drift then CTRL + Z and clicking in the Track Editor causes a crash |
| 9118 | Annotate area can be activated even when licence is disabled |
| 9115 | Voxel Calibration of TIFF exported by ImageJ 1.47b (Fiji) not read |
| 9105 | Track Editor numbering doesn't match the frame number when analysing a subset of data |
| 9103 | 3D cursor should disappear when loading new image |
| 9086 | Merging Spots objects then rebuilding tracks stalls Imaris |
| 9085 | Changing the displayed statistics while in the Colour tab will lead to multiple instances of Points and Tracks displaying |
| 9073 | 3D View flickers at some zoom levels with files |
| 9013 | Vesical histogram disappear after swithing tabs |
| 8994 | ZVI file crashes Imaris with 'Out of Memory' Message |
| 8993 | Cannot read compressed ZVI files |
| 8991 | Imaris is really slow if Arena has many batch runs |
| 8961 | Show Help from the context menu for labels leads to a page that does not exist |
| 8952 | Missing labels columns in statistics tab after adding more statistics type through preferences |
| 8940 | Cells parameters are altered in duplicate |
| 8933 | Colour Type in the material editor should revert to 'Base' on rebuild |
| 8916 | Explain the Arena Preference options when moving the database in detail |
| 8896 | Spines are branched even though that function was unchecked, during the creation of the filament. |
| 8886 | Link Vesicle Track-ID's with Cell Track-ID's |
| 8860 | Selecting specific statistics for Filament is still showing all filament stats |
| 8846 | Filename in Fiji is not the filesystem name |
| 8797 | The time column in Export Data For Plotting is incorrect |
| 8783 | Timestamps from LIF files not read correctly (from new LAX software) |
| 8780 | File series detection limited to '999' files |
| 8762 | Launch of AutoQuant from 'Image Processing - AutoQuant' fails |
| 8718 | PlotNumbersArea Doubleclick to open original image does not work for Collections of 1 item |
| 8701 | Mac does not support credentials that require a workgroup |
| 8696 | ROI in Surfaces is very small and not in the center anymore |
| 8694 | Various Missing links |
| 8680 | Imaris incorrectly warns the user after opening the file using 'image only' even after using store as |
| 8652 | Cannot access 'Edit Spots' options (stylesheet) |
| 8620 | Coloc parameter naming for ROI usage is inconsistent |
| 8550 | All Spine Part Length(s) Incorrect (many negative and zero values) |
| 8466 | Auto-path won't add dendrites even signals are strong |
| 8370 | Crash when stored creation parameters are used |
| 8295 | Not all z slices are read in for an tiff file |
| 8028 | Imaris does not read Nikon Elements ND2 Time interval (or reads incorrect time) |
| 7850 | Filters in Wizard 'ignored' delivering false results |
| 7796 | Missing texture blocks after Resampling Open (.lif) |
| 7681 | Nucleus transparency has no effect |
| 7633 | Import Spots and Surfaces to Cells does not work with mismatching times |
| 7613 | Imaris keeps file read locks on some loaded tiff images |
| 7530 | Free rotate does not allow normal editing of Axis values |
| 7528 | Disabling the currently selected statistics colour coding type can cause Imaris to swap to a different surpass object |
| 7449 | Edit single time point calibration |
| 7423 | Big Tiff files loading only first frame - not entire image. |
| 7189 | Volume statistics tab not updated when switching from Surface object |
| 7183 | Cellbody segmentation - split by seed points does not split as expected |
| 7152 | contour tool can not create surface if voxel size is small |
| 7114 | Update compiled Xtensions to newest Matlab version |
| 7090 | Imaris fail to open .cxd file if data contain z information |
| 6889 | Multiscale Watershed |
| 5979 | Duplicate Selection should also duplicate the creation parameters |
| 5868 | Make it apparent to the user if he is using normal matlab or runtime |
| 5692 | split cells by seed point gives fewer cells than seed points |
| 5330 | ImarisCell continues to miss these cells that are in close proximity but clearly different cells. |
| 5281 | Find location of installed Matlab and Matlab runtime on MAC |
| 5046 | Camera Drift Correction (without resampling the image) |
| 4641 | Changes to image composition (i.e. adding or deleting channels), all channels become visible |
| 4508 | statistics channels not updated after delete channels |
| 4030 | ND2 file reader problem: timelapse with multiple XY positions not opened properly |
| 2740 | ImageJ Tiff can not be read |